 TissUUmaps Hackathon, 2021-09-29
TissUUmaps Hackathon, 2021-09-29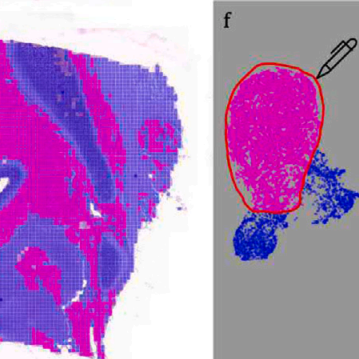
DEPICTER: Deep representation clustering for histology annotation.
Computers in Biology and Medicine, doi: 10.1016/j.compbiomed.2024.108026, January 26. 2024.
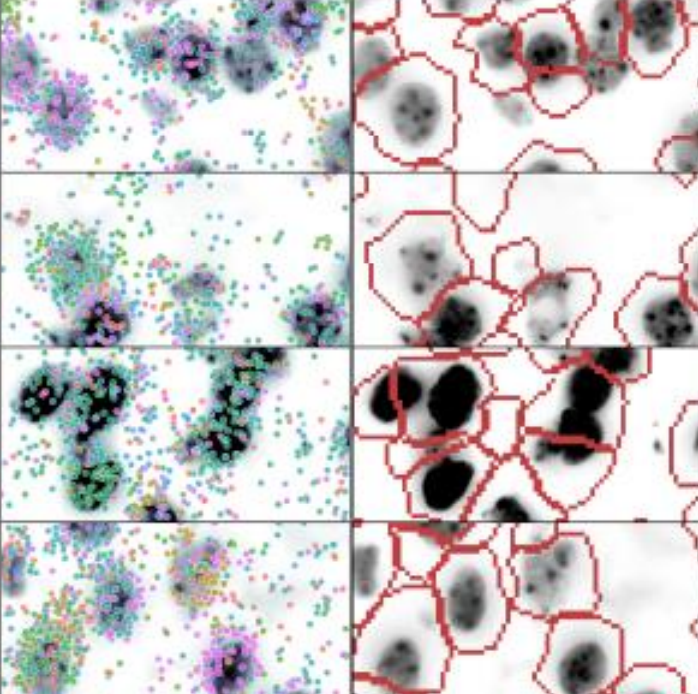
Cell segmentation of in situ transcriptomics data using signed graph partitioning.
Lecture Notes in Computer Science, Presented at the International Workshop on Graph-Based Representations in Pattern Recognition, GbRPR 2023, doi: 10.1007/978-3-031-42795-4_13, August 24. 2023.
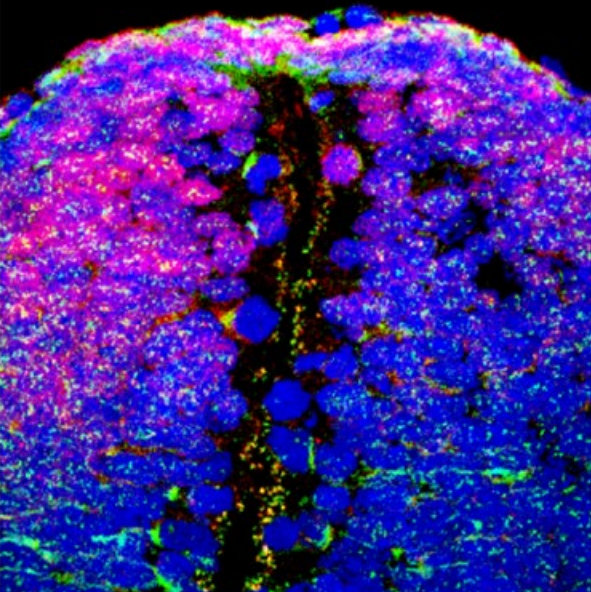
Profiling spatiotemporal gene expression of the developing human spinal cord and implications for ependymoma origin.
Nat Neurosci, doi: 10.1038/s41593-023-01312-9, April 24. 2023.
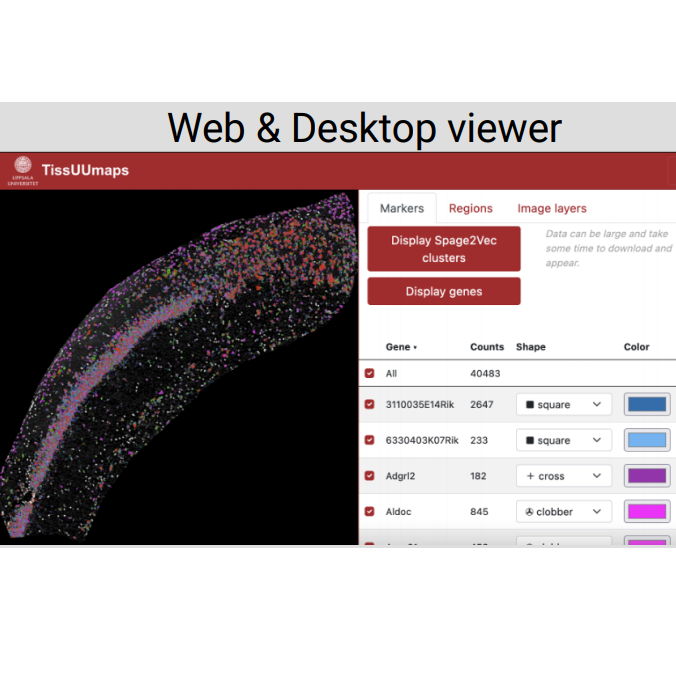
TissUUmaps 3: Interactive visualization and quality assessment of large-scale spatial omics data.
Heliyon, doi: 10.1016/j.heliyon.2023.e15306, April 21. 2023.
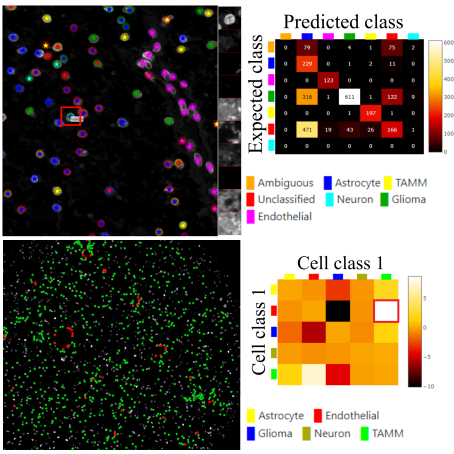
Visualization & Quality Control Tools for Large-scale Multiplex Tissue Analysis in TissUUmaps 3.
Biological Imaging, doi: 10.1017/S2633903X23000053, February 20. 2023.
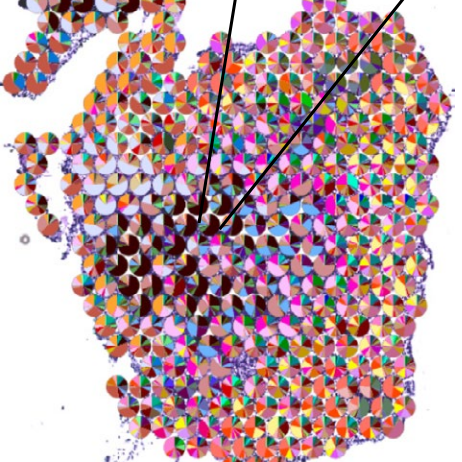
A topographic atlas defines developmental origins of cell heterogeneity in the human embryonic lung.
Nat Cell Biol, doi: 10.1038/s41556-022-01064-x, January 16. 2023.
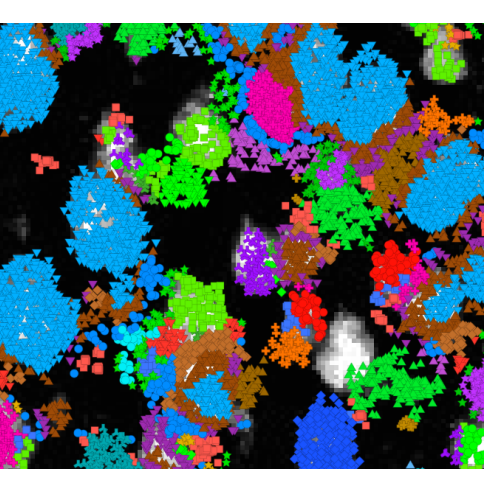
Points2Regions: Fast Interactive Clustering of in Situ Transcriptomics Data.
bioRxiv, doi: 10.1101/2022.12.07.519086, December 10. 2022.
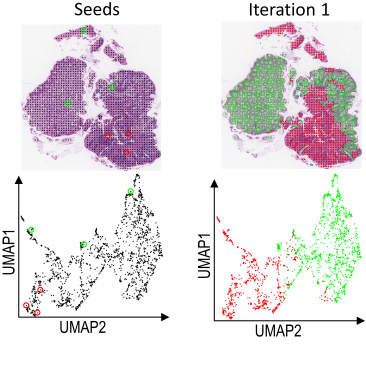
Seeded iterative clustering for histology region identification.
arXiv, doi: 10.48550/arXiv.2211.07425, November 14. 2022.
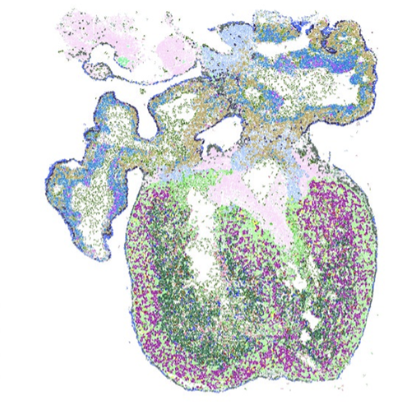
De novo spatiotemporal modelling of cell-type signatures in the developmental human heart using graph convolutional neural networks.
PLoS Comput Biol, doi: 10.1371/journal.pcbi.1010366, August 12. 2022.
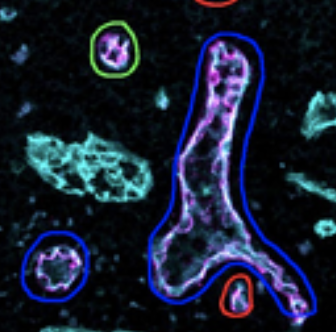
Automated detection of vascular remodeling in tumor-draining lymph nodes by the deep-learning tool HEV-finder.
Journal of Pathology, doi: 10.1002/path.5981, June 13. 2022.
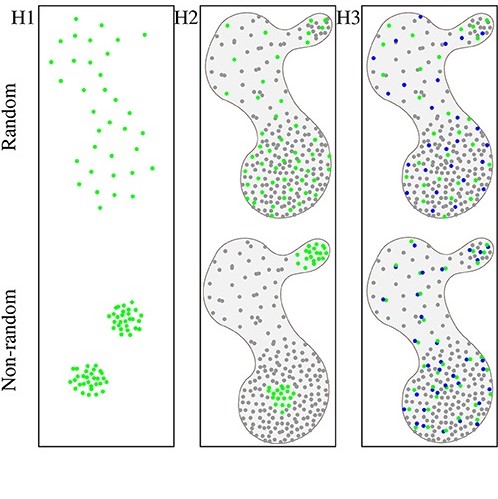
Spatial Statistics for Understanding Tissue Organization.
Frontiers in Physiology, doi: 10.3389/fphys.2022.832417, January 28. 2022.
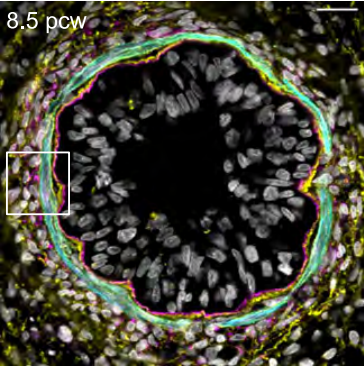
Developmental origins of cell heterogeneity in the human lung.
bioRxiv, doi: 10.1101/2022.01.11.475631, January 12. 2022.
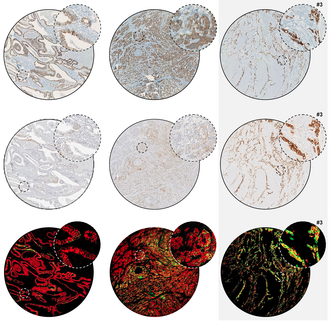
Comparison of East-Asia and West-Europe cohorts explains disparities in survival outcomes and highlights predictive biomarkers of early gastric cancer aggressiveness.
Int J Cancer, doi: 10.1002/ijc.33872, Dec 2, 2021.
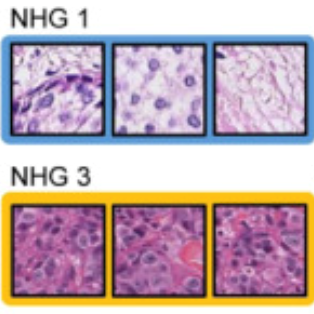
Improved breast cancer histological grading using deep learning.
Annals of Oncology, doi: 10.1016/j.annonc.2021.09.007, Sept 29. 2021.
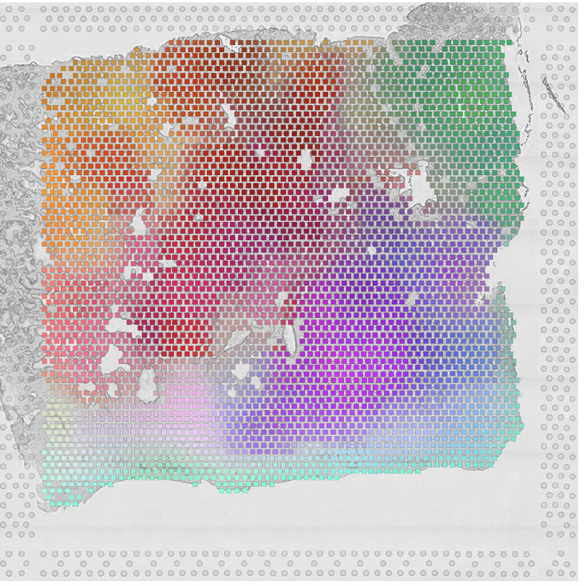
Morphological Features Extracted by AI Associated with Spatial Transcriptomics in Prostate Cancer.
Cancers, doi: 10.3390/cancers13194837, Sept 21. 2021.
Artificial Intelligence for Diagnosis and Gleason Grading of Prostate Cancer in Biopsies - Current Status and Next Steps.
European Urology Focus 7(4) p687-691, doi: 10.1016/j.euf.2021.07.002, July 2021.
Machine learning for cell classification and neighborhood analysis in glioma tissue.
Cytometry, doi: 10.1002/cyto.a.24467, June 4. 2021.
ISTDECO: In Situ Transcriptomics Decoding by Deconvolution.
doi: https://doi.org/10.1101/2021.03.01.433040, March 02, 2021.
CoMIR: Contrastive Multimodal Image Representation for Registration.
In proceedings of Neural Information Processing Systems 2020 (NeurIPS 2020), Nov. 2020.

Automated identification of the mouse brain’s spatial compartments from in situ sequencing data.
BMC Biology, doi.org/10.1186/s12915-020-00874-5, Oct 2020.
Spage2vec: Unsupervised representation of localized spatial gene expression signatures
FEBS Journal, doi: 10.1111/febs.15572, Sept 2020.
Regular use of depot medroxyprogesterone acetate causes thinning of the superficial lining and apical distribution of HIV target cells in the human ectocervix.
The Journal of Infectious Diseases, doi: 10.1093/infdis/jiaa514 Aug 2020.
Graph-based image decoding for multiplexed in situ RNA detection.
To appear in the proceedings of the International Conference on Pattern Recognition (ICPR), 2020. A pre-publication with similar content can be found here; Permanent arXiv identifier: 1802.08894.
Towards automatic protein co-expression quantification in immunohistochemical TMA slides.
IEEE Journal of Biomedical and Health Informatics, doi:10.1109/JBHI.2020.3008821, July 2020.
Deep learning and conformal prediction for hierarchical analysis of large-scale whole-slide tissue images
IEEE Journal of Biomedical and Health Informatics, doi:10.1109/JBHI.2020.2996300, June 2020.
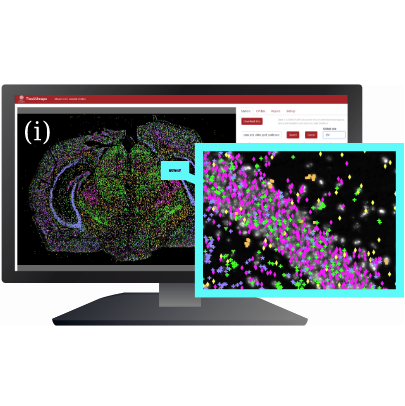
TissUUmaps: Interactive visualization of large-scale spatial gene expression and tissue morphology data.
Bioinformatics, doi:10.1093/bioinformatics/btaa541, May 2020.
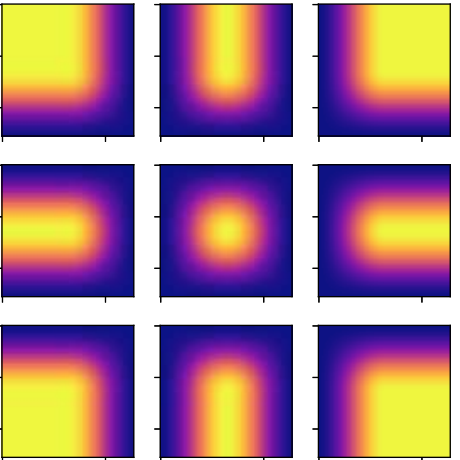
Introducing Hann windows for reducing edge-effects in patch-based image segmentation.
PLoS One 15(3):e0229839. Mar 12, 2020, doi:10.1371/journal.pone.0229839.
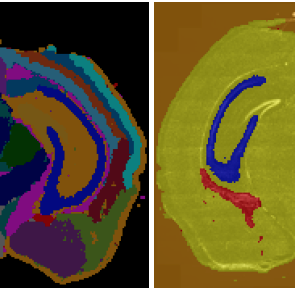
Transcriptome-supervised classification of tissue morphology using deep learning
Proc IEEE Int Symp Biomed Imaging 2020, April 2020, doi:10.1109/ISBI45749.2020.9098361
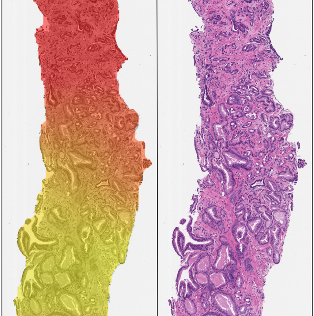
Artificial intelligence for diagnosis and grading of prostate cancer in biopsies: a population-based, diagnostic study.
Lancet Oncology, doi: 10.1016/S1470-2045(19)30738-7, Jan 8, 2020. The paper is mentioned in Läkartidningen.

Impact of Q-Griffithsin anti-HIV microbicide gel in non-human primates: In situ analyses of epithelial and immune cell markers in rectal mucosa.
Scientific Reports, doi: 10.1038/s41598-019-54493-4 Dec 2, 2019.
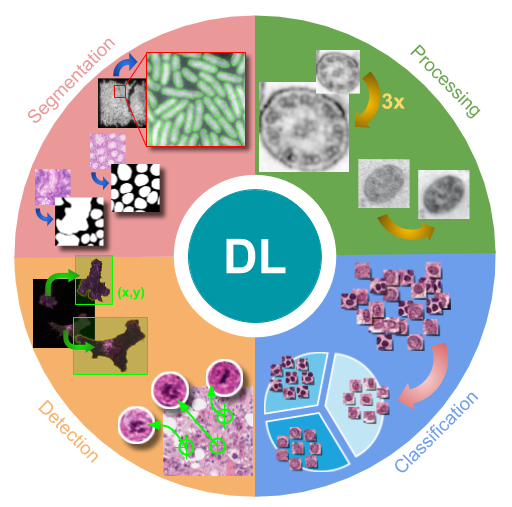
Deep Learning in Image Cytometry: A Review.
Cytometry A. doi: 10.1002/cyto.a.23701, Dec 19 2018.
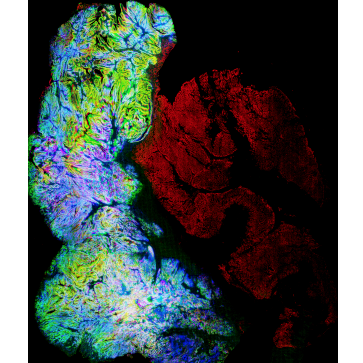
Whole Slide Image Registration for the Study of Tumor Heterogeneity.
Presented at COMPAY workshop of MICCAI 2018 21st International Conference on Medical Image Computing and Computer Assisted Intervention, September 16-20 2018, Granada, Spain, published in Lecture Notes in Computer Science, Computational Pathology and Ophthalmic Medical Image Analysis; LNCS 11039, pp 95-102, Springer, Cham. doi: 10.1007/978-3-030-00949-6_12

Decoding Gene Expression in 2D and 3D.
Presented at SCIA17 (Scandinavian Conference on Image Analysis), Tromsö, Norway, June 12-14, 2017, published in Lecture Notes in Computer Science; LNCS 10270: 257-268. Springer, Cham.

Bridging Histology and Bioinformatics - Computational Analysis of Spatially Resolved Transcriptomics.
Proceedings of the IEEE 99, doi: 10.1109/JPROC.2016.2538562, April 2016.
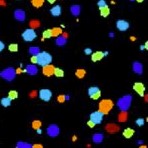
Image based in situ sequencing for RNA analysis in tissue.
Proc IEEE ISBI 2014, International Society of Biomedical Imaging, 29 April - 2 May, 2014, Beijing, China.
In situ sequencing for RNA analysis in preserved tissue and cells.
Nature Methods, 2013 (10), 857-860. doi: 10.1038/nmeth.2563. PMID: 23852452
The project is funded by the European Research Council, through an ERC consolidator grant to Carolina Wählby (ERC CoG 682810).
Important support was also provided via the Swedish Foundation for Strategic Research (SSF), via the SysMic project (SB16-0046), with Staffan Strömblad as PI and Carolina Wählby as co-PI.
Some of the activities are also funded by SSF Big Data grant on ‘Hierarchical Analysis of Temporal and Spatial Image Data - From intelligent data acquisition via smart data-management to confident predictions’, where Carolina Wählby acts as PI.
TissUUmaps is also supported as a technology development project which is part of the SciLifeLab BioImageInformatics Facility of SciLifeLab, a national infrastructure for life science research in Sweden.


